 |
| Early Warning’s Pathogen Sensor offers a new performance curve for pathogen detection by automating labor-intensive and decades-old technologies to provide:
- Higher likelihood of detecting infectious pathogens
- Shortened sample collection and test cycle from 2 -14 days down to 3 hours
- Reduced cost per test by 70% to 90%
|
.gif) |
Testing is the most effective way to prevent the transmission of pathogens, as pathogen testing of representative samples can initiate immediate actions to stop the spread. The performance of biotesting technologies can be measured on the following principal parameters:
1. Likelihood of detection pathogens - based capturing sufficient pathogens from a localized event and then distinguishing actual pathogens from harmless microorganisms. The ability to successfully detect pathogens is impacted by:
- Sample Size – if the sample is too small doesn’t capture the pathogen the negative test result is misleading as a larger sample size would have a greater chance of capturing a minimal amount of pathogen that could be detected
- Test Frequency – if the sample isn’t tested when the pathogen event occurs, the negative test result is again misleading as more frequent testing would have a greater the chance of capturing the pathogen
- Test Sensitivity – the more sensitive the test, the greater the ability to distinguish a signal from the pathogens reacting with the biosensor from background noise
- Test Specificity – the more specific the test, the greater the ability to distinguish specific target pathogens from non-specific microorganisms and other materials
2. Sample-to-Report Time - where the faster the pathogen is detected the greater the chance that the contamination can be stopped before pathogens reach consumers or valuable products. The ability to rapidly detect pathogens is impacted by:
- Time for collecting a sample – including travel to and from a remote location
- Time for each test process - the sum of the individual processes to time to amplify, detect, interpret and report the results
- Wait time between test processes – Any backlog in the laboratory, downtime from repairs, or unavailability of technicians will add to the test time
- Time for repeating tests - a positive or suspicious result requires confirmation from subsequent testing and further delays to ensure that the initial result was valid
3. Test Cost - as the high cost per test limits the number of tests that can be undertaken by government agencies, commercial organizations, and consumers due to budget constraints. Test costs are impacted by:
- Direct Test Costs – including laboratory costs and consumables, interpret test results, and reporting to end users
- Sampling Cost – including labor, travel and transport
- Number of Pathogens per Test – which may be increased for a suite a potential pathogens
- Cost for repeating tests - a positive or suspicious result requires confirmation from subsequent testing and further delays to ensure that the initial result was valid.
Pathogen tests and their associated collection methods vary in their effectiveness in the following ways:
Laboratory Pathogen Tests (e.g. Immunoassay, Molecular PCR) - are currently done in government-certified biosafety laboratories using expensive equipment and highly trained technicians who oversee pathogen tests under highly stringent conditions. Current technologies include molecular polymerase chain reaction (PCR) and immunoassays which are processed in batches for detecting a single pathogen. Pathogen tests can take several days or weeks to analyze a suite of suspected pathogens. Each sample has to be manually collected and delivered to the lab without contamination which adds considerable cost and time to the testing procedure. Because of the limitations in transporting multiple samples, the standard sample size is only 100 mLs. This small sample size has a very low probability of capturing pathogens as they are unevenly distributed in the source.
Laboratory Screening Tests (e.g. Enzyme Culture) - An alternative to laboratory pathogen tests is laboratory screening tests that culture cells of indicator organisms such as Coliforms or indicator E.coli. Screening tests are cheaper, easier to administer, and generate results more quickly than pathogen tests. However, most of the microorganisms that culture in a screening test are harmless bacteria, which would generate a positive test outcome and necessitate subsequent pathogen tests to more effectively validate the suspicious result. This two step process adds days to total detection time. Coliforms and indicator E.coli cultures are also limited in scope as they do not detect the presence of parasites, viruses and non-fecal bacteria which make up the majority of the waterborne pathogens listed by the World Health Organization. Screening culture tests also need to be conducted in biosafety laboratories and require manual sampling of 100 mL samples which adds to the cost and further reduces the likelihood of detecting pathogens.
Inline Pre-screening Instruments - A third approach makes use of pre-screening instruments that measure indirect parameters that could potentially be associated with microorganisms. While these measurements can be instantaneous, none of these parameters distinguish pathogens from harmless comparative materials that can outnumber pathogens by millions to 1, such as heterotrophic bacteria, organic materials, plankton, fibers, chemicals, rust, and dust. This makes their usefulness limited for pathogen detection. Pre-screening instruments include:
- Turbidity Meters – that measure turbidity as the degree of cloudiness of water or liquid from all materials in a sample which may or may not include pathogens
- Particle Size Analyzers - that measure the distribution of particles by size from all materials in the sample which may or may not include pathogens
- Biological Oxygen Demand (BOD) Analyzers - which measure dissolved oxygen from all living organisms in the sample which may or may not include viable pathogens
- Adenosine Triphosphate (ATP) Luminometers - which measure residual amounts of the chemical marker adenosine triphosphate (ATP), a compound found in all types of plant, animal and microbial cells, which may or may not include pathogens
Inline Pathogen Sensor - Early Warning’s inline pathogen sensor has both a high likelihood of detecting pathogens and a short sample to report time to bring pathogen detection to a new performance curve. Early Warning employs a revolutionary nanotechnology-based biosensing platform exclusively licensed from NASA that can detect 10 or more specific pathogens per test rather than a single pathogen or indicator organism. Detection performance and processing time are further improved with an automatic sampler and pathogen concentrator. This allows a 10 Liter sample to be processed and provides a 100 times greater chance of capturing pathogens than a standard 100 mL sample. The fully automated pathogen sensor provides sample to results in 3 hours instead of 2 to 14 days. By avoiding costly manual sampling, expensive biosafety laboratories, and highly trained technicians, the cost per test is reduced by 70 to 90%.
Early Warning’s pathogen sensor provides a high likelihood of detecting pathogens, rapid sample-to-report time, and low cost per test from the following overall capabilities:
Sensitivity – The ability to detect very low levels of target materials using:
- Small sensor area for working electrodes and corresponding low signal-to-noise ratio
- 18-bit digital potentiostat capable of distinguishing low resolution signals at pico amps
- Concentrating 10 Liter samples which comprise 100 times more pathogens than a traditional 100 ml sample and targeting RNA which has 20,000 times more target materials for detection than a single cell making 1,000,000 times as many detection targets (factoring in yield) than a traditional cell analysis
Specificity – The ability to distinguish between different target pathogens using:
- Individual working electrodes with commercial DNA probes that only bind with 30 or more base pairs in matching RNA
- Removal of potentially interfering materials using magnetic bead separation that extract target organisms for detection and leaving behind heterotrophic bacteria and other non specific materials
Accuracy – The ability to correctly measure the concentration of target materials through:
- Sampling a bigger water volume than the traditional grab sample which has a higher chance of capturing the portion of water containing the pathogen
- Having the ability to measure frequently which has a higher chance of capturing the portion of water containing the pathogen
- Directly measuring the pathogenic materials which could be missed by an indicator test
Multiplexing – The ability to measure multiple pathogens simultaneously using:
- Small sensor area for working electrodes that allows many to fit on a very small surface
- Multiplexing potentiostat that can measure current signals from multiple channels at the same time
An ultra-sensitive DNA-Electrochemical Biosensor tests for multiple pathogens at the same time
|
Early Warning’s Biosensor is an analog biochip that directly measures individual species of pathogenic bacteria, protozoa and viruses in the same test. The biochip contains nine or more independent working electrodes connected to separate channels of a digital potentiostat, along with a counter electrode and reference electrode in a water-tight casing. Each working electrode has a different set of highly specific DNA probes associated with a unique target pathogen, such as E.coli O157:H7 or Salmonella. When single strands of RNA from the sample come into contact with DNA probes, complementary strands hybridize into double helixes. Only exact matches bind to ensure high specificity of the test results. The potentiostat applies a voltage scan which generates electrical current from guanine oxidation only at the working electrodes where target RNA hybridizes. The peaks of the current are measured against known samples to provide a concentration level of target pathogens that are present in the sample. To avoid contamination, each biosensor chip is used once. A number of biochips are packaged in a replacement cartridge.
|
|
|
Optimum Core Technologies
Early Warning’s DNA Electrochemical Biosensor employ core components that are Very Specific, Ultra-sensitive and Low Cost.
|
A biosensor comprises two main components:
1. Bioreceptor - which is a biological mediator that causes a target analyte to be recognized. Common bioreceptors include:
-
Antibody – which attract to antigens
- Enzyme – which increase the rate of a chemical reaction, and
-
Nucleic Acid – which form base pairs with complementary nucleic acids strands Nucleic Acid bioreceptors can be very specific by providing a long sequence of bases that hybridize only with the biomolecules being targeted.
2. Transducer - which converts the bioreceptor-analyte interaction into a measurable signal. Common transducers include:
-
Optical – which measures the change in optical properties such as color, light output or absorbance
-
Electrochemical – which measures the change in electrical properties such as electrical current or electrical potential
Electrochemical transducers can be ultra-sensitive and low cost by adapting scalable techniques used in semiconductor fabrication. Early Warning’s Pathogen Sensor employs core components that are very specific (Nucleic acid), ultra-sensitive and low cost (Electrochemical).
|
Breakthrough Working Electrode Design
Enables nucleic acid bioreceptor probes (bioprobes) using a tiny footprint to fit multiple working electrodes on the same sensor for multiplexing. 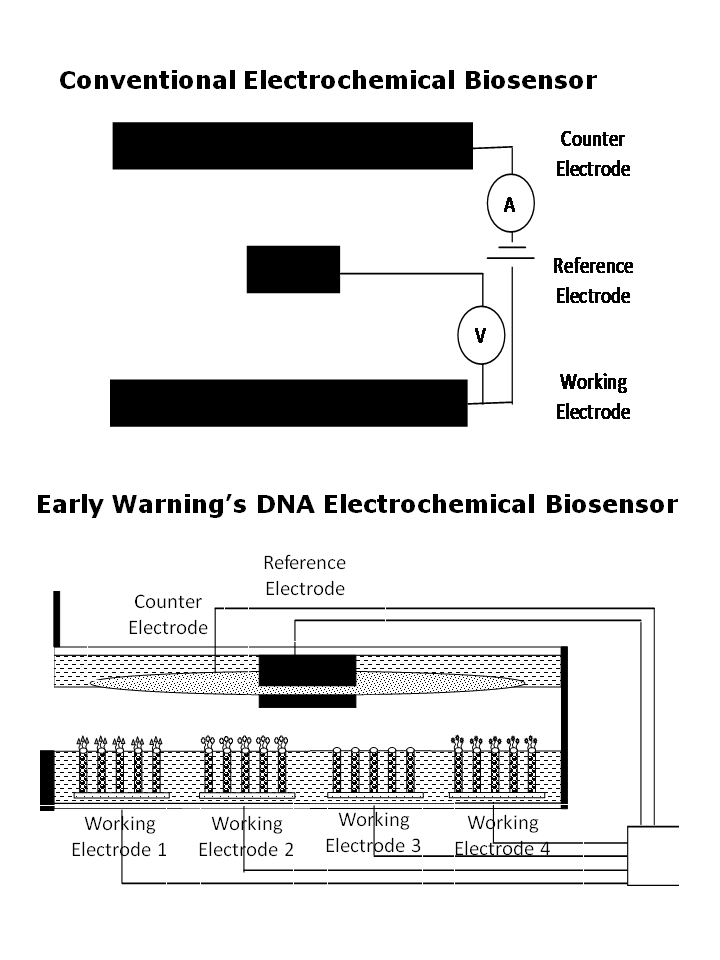
|
Electrochemical sensors use three electrodes:
-
Working electrode where a target material either loses electrons through oxidation or gains electrons through reduction,
-
Counter electrode that completes an electrical circuit and creates an electrical current by having the generated electrons flow into and then leave the solution via the working electrode and the counter electrode, and
-
Reference electrode which compensates for any change in the electrical conductivity caused by the reaction and provides a reference for measuring voltage.
Most electrochemical sensors detect a single chemical, metal or material since different materials oxide or reduce at different applied voltages and generated currents. In the case of nucleic acid bioreceptors, guanine is a natural constituent of DNA and RNA and oxidizes at 1.05 volts for all types of biomolecules, and can accommodate the detection of multiple biomolecules using an electrochemical transducer.
Early Warning’s pathogen sensor takes advantage of guanine’s activity at 1.05 volts, and employs 9 or more working electrodes on the same biochip. Each working electrode has a base pad with perpendicular nanowires and a pathogen-specific set of DNA probes attached at the nanowire tips. Because of their small size, many working electrodes can be placed in parallel on the same biochip to test for multiple pathogens at the same time.
|
|
Highly Specific Detection of Individual Species
DNA probes of specific pathogens hybridize with matching rRNA from target pathogens. When voltage is applied only exact matches form double helixes and generate current from guanine oxidation. 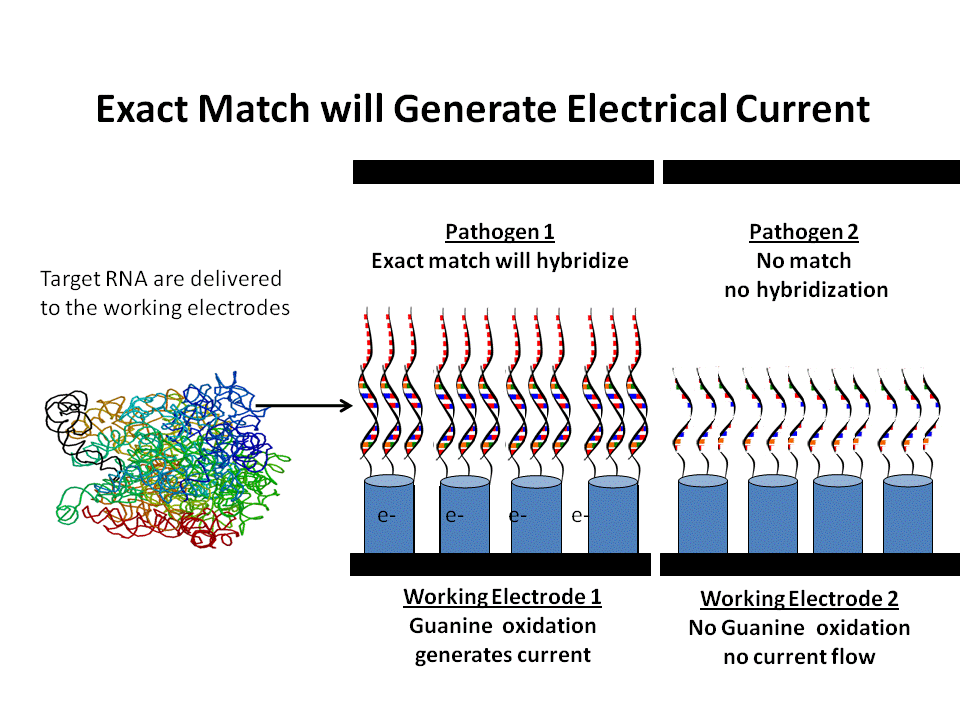
|
Each target pathogen has its own working electrode which contains DNA bioprobes chemically attached to the tips of nanowires. The DNA probes contain 30 or more base sequences that make the probes extremely specific to the rRNA of a specific pathogen or individual species such as E.coli O157:H7.
The single strands of RNA from the sample come into contact with the bioprobes and only complementary strands hybridize into double helixes. When 1.05 volts are applied across the electrodes, guanine reacts with cytosine only where hybridization takes place from an exact match. This causes a flow of electrons to the designated working electrode. The magnitude of the electrical current is related to the amount of rRNA in the sample and subsequently to the concentration of the pathogen in the sample. If there is no hybridization from the absence of the target pathogen, there is no current.
|
Ultra-sensitive detection of low dose pathogens
From an ultra-small precisely ordered working electrode surface that can distinguish nanoAmp signals from background noise.
|
Early Warning’s pathogen sensor distinguishes current signals from guanine oxidation at very low pathogen concentration from background noise by employing an ultra-small surface area on its working electrodes. The area of the working electrode surface in an Early Warning sensor is 0.000003 cm2 which is approximately 5 orders of magnitude smaller than the working electrode in a commercial glucose monitor.
Early Warning’s working electrodes have highly conductive nanowires extending perpendicularly from the working electrode base. The nanowires are made from highly ordered carbon nanofibers that form a small surface at the nanowire tips. The nanowires are separated by electrical insulating material and have DNA probes attached at the tops. Nanofabrication methods permit the nanowires to be spaced at least 1 micron apart to avoid touching and subsequently prevent false signals. Currents generated from Early Warning’s working electrode are in the order of 1 – 10 nanoAmps which are measured by an onboard 18-bit digital potentiostat capable of distinguishing low resolution picoAmp signals from each working electrode.
|
Electrochemical Detection
The magnitude of the peak current can be converted into a concentration level using pre-determined values.
|
When guanine oxidizes after hybridization takes place the flow of elections is extremely low making the resulting electrical current difficult to measure. A Ruthenium bipyridine mediator which also oxidizes at 1.05 volts can amplify the signal by transferring guanine electrons to the electrode surface and improving the measurement accuracy. In this approach Ruthenium bipyridine Ru(bpy) is used to oxidize guanine following electrochemical activation as follows:
.gif)
Both guanine and Ruthenium bipyridine lose electrons though oxidization when electrical potential voltage is applied. When the voltage scan is reversed, Ruthenium bipyridine will regain its electrons but guanine will not. As a Ruthenium bipyridine can oxidize and reduce repeatedly and form a baseline to measure the incremental current from guanine alone. The electrical current from Guanine and Ruthenium bipyridine combined in a first scan is much greater than Guanine alone which needs to be distinghushed from the background noise for the presence/absence detection threshold.
The following curves show the results from 3 consecutive oxidation scans where:
Scan 1: is from E.coli’s Guanine oxidation PLUS Ru(bpy) mediator oxidation which also transports guanine electrons to the electrode surface
Scan 2: Is from only the Ru(bpy) mediator oxidation. The difference of peak current from Scan 1 minus Scan 2 is from E.coli. The greater the E.coli concentration, the greater the current.
Scan 3: Is from only the Ru(bpy) mediator oxidation. The difference of peak current from Scan 2 minus Scan 3 is background noise that determines the threshold value for the E.coli signal which is the variability or maximum difference in two scans of Ru(bpy). The current from background noise must be exceeded for E.coli to be present.
|
An automated Pathogen Concentrator eliminates the need for manual sampling and PCR amplification
|
Early Warning’s Pathogen Concentrator greatly reduces the time involved in sampling and preparing pathogens prior to detection by placing a biosafety laboratory in an automated inline instrument. Samples do not have to be transported to the lab and as a result the 100 mL sample size is no longer a constraint. By increasing the sample size from 100 mL to 10,000 mL (10 L), 100 times more pathogens are collected in a composite sample, and there is a 100 times higher chance of capturing pathogens in a skewed distribution. The larget sample also has a much higher chance of capturing highly infectious protozoa and viruses typically found in very low concentrations. Samples are further processed to extract approximately 20,000 RNA per cell. The equivalent of 1 cell per 100 mL sample translates to millions of RNA targets from a 10 Liter sample which is sufficient material for the biosensor to detect pathogens without PCR or amplification. Once captured and concentrated, target pathogens are sent for detection to an unused biochip. The sample-to-report time is about 3 hours. Test results are transmitted by the concentrator through wired or wireless communications to allow preventative measures to be taken before contaminated batches reach valuable products or consumers.
|
|
Filtration
Filtration Ultrafiltration removes filtered water while concentrating viruses, bacteria, protozoa, clumps and biofilms.
|
The 10 Liter sample is automatically collected and moved through a pressure regulator, and flow meter to a reservoir. From the reservoir, the sample is fed through a semi-permeable ultrafiltration membrane. A 50 kilodalton membrane with a pore size of about 15 nm diameter allows small water molecules to pass while retaining viruses (15 - 30 nm), bacteria (100 – 5,000 nm) and protozoa (4,000 nm and higher) in a retenate. The retentate is continuously filtered while ultra clean filtrate water, free of microorganisms, is pumped away to a separate reservoir. This leaves a highly concentrated retentate of 200 milliliters loaded with potential target pathogens. This process is then repeated through a smaller ultrafiltration membrane to further concentrate the retentate down to 10 milliliters. Various chemicals, additives and filters are used to remove or neutralize chlorine, large particles and other materials that could potentially interfere with detecting target pathogens. A process recipe can be pre-selected or fine-tuned depending on the complexity of the input water or liquid.
|
Pathogen Separation
Magnetic beads with antibodies attach to target pathogens so they can be extracted from non-specific biomaterials.
|
Immunomagnetic separation is used to extract pathogens from heterotrophic bacteria and other harmless microorganisms. Iron core beads coated with antibodies are provided to attract target pathogens to their surface. When a magnet is activated, drawing the beads to it, the heterotrophic bacteria and non-specific materials are washed away. The magnetic bead retentate containing only the target pathogens are condensed to about 1 milliliter which is moved into the microfluidics portion of the Concentrator.
|
RNA Extraction
Microfluidics are used to prepare RNA from target pathogens and deliver to an available biosensor for pathogen detection
|
The retentate from the immunomagnetic separation contains pathogens attached to magnetic beads. The retentate undergoes lysozyme, which destroys the cell walls of the target pathogens releasing the cell contents. All DNA is destroyed with a DNAse enzyme and the sample then passes through a silica filter, allowing only the ribosomal RNA to pass through. The single strand RNA are delivered to an unused biochip to allow hybridization with complementary DNA probes, and then electrochemical detection.
The biosensor detects RNA from all pathogens (viable and non-viable). An optional Viable Pathogen Test can be used to distinguish viable cells from total cells. In viability testing, the sample is metered into two separate 500 microliter samples. The first, or “Total Pathogen Test”, undergoes lysozyme, DNAse processing, and rRNA separation which is sent to an unused biochip for pathogen detection. The second sample, or “Viable Pathogen Test”, is first fed specific nutrients, heated to 37 degrees Celsius and mixed to allow viable cells to rejuvenate and reproduce for about one and a half hours. It then undergoes lysozyme treatment. RNA in the nutrient treated sample, or “Viable Pathogen Test”, are detected in a similar manner on a second biochip. If the viable cells are present and reproducing, the increased amount of ribosomal RNA will produce a higher current which allows for an accurate representation of the viability of the first, or “Total Pathogen Test". This increase in current is calibrated with known samples to provide an estimate of the percent viability for each pathogen.
|
Transmit Test Results
Onboard communications support RS232, wired Ethernet and WiFi
|
Test results are reported on a user interface screen that can be viewed locally using a laptop or remotely using a SCADA software module that can be interfaced with a user’s existing SCADA or control system. The Concentrator’s User Interface provides progress bars of each process stage and generates an audit trail of each process that is successfully undertaken. In the event that an alert level is reached, the Concentrator sends a Biohazard Alert message to preprogrammed destinations. These can include SCADA screens or PDAs. Once the test is completed the system automatically sanitizes itself using a sanitizing agent mixed with the filtered water. After 60 minutes the system is flushed and ready for a new test.
|
|
|